Each nucleosome has its own physical properties - not just because of differences
in the protein core but especially as a result of the sequence dependent shape and
mechanics of the wrapped DNA double helix. We have studied in detail how this
affects the dynamic properties of nucleosomes. One example is nucleosome breathing:
parts of the wrapped DNA unwrap spontaneously from the protein core. We use our
coarse grained nucleosome model to interpret various types of
experiments (restriction enzymes, FRET, SAXS) and to highlight the differences between nucleosomes
containing different base-pair sequences
(Culkin 2017; van Deelen 2020).
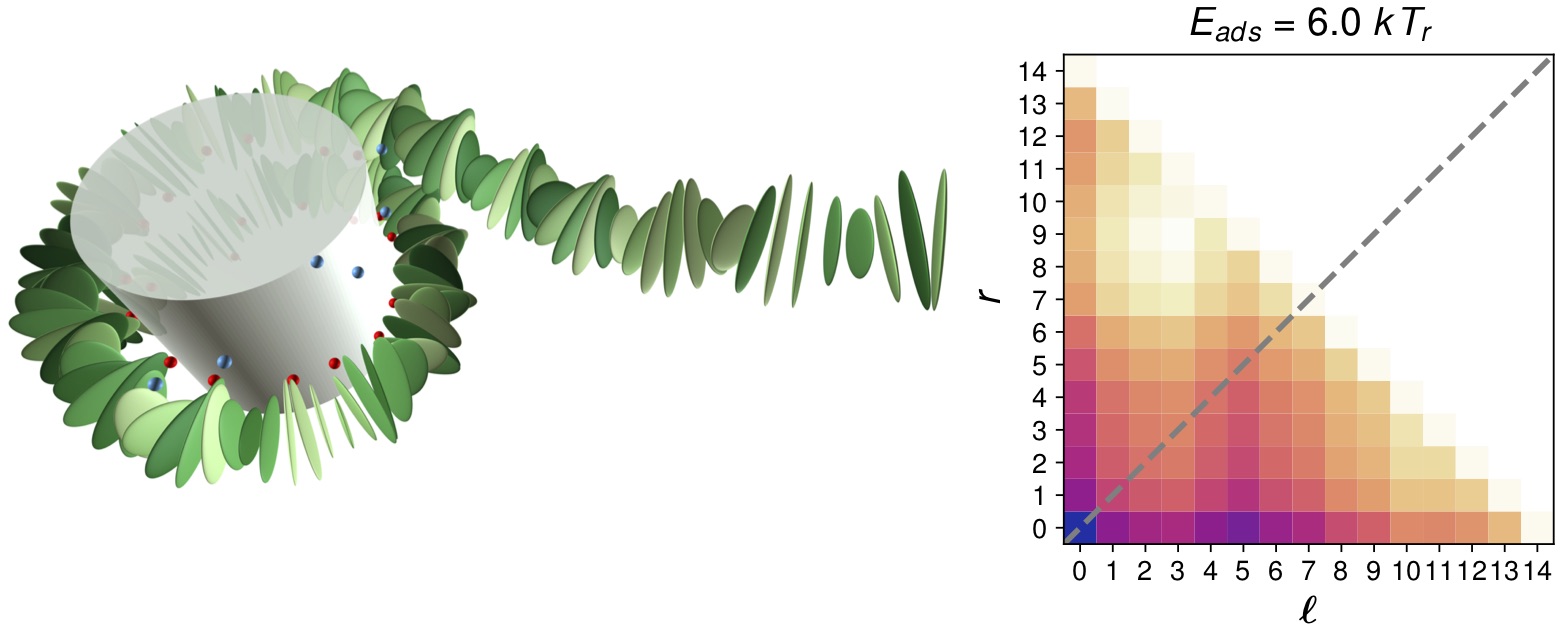
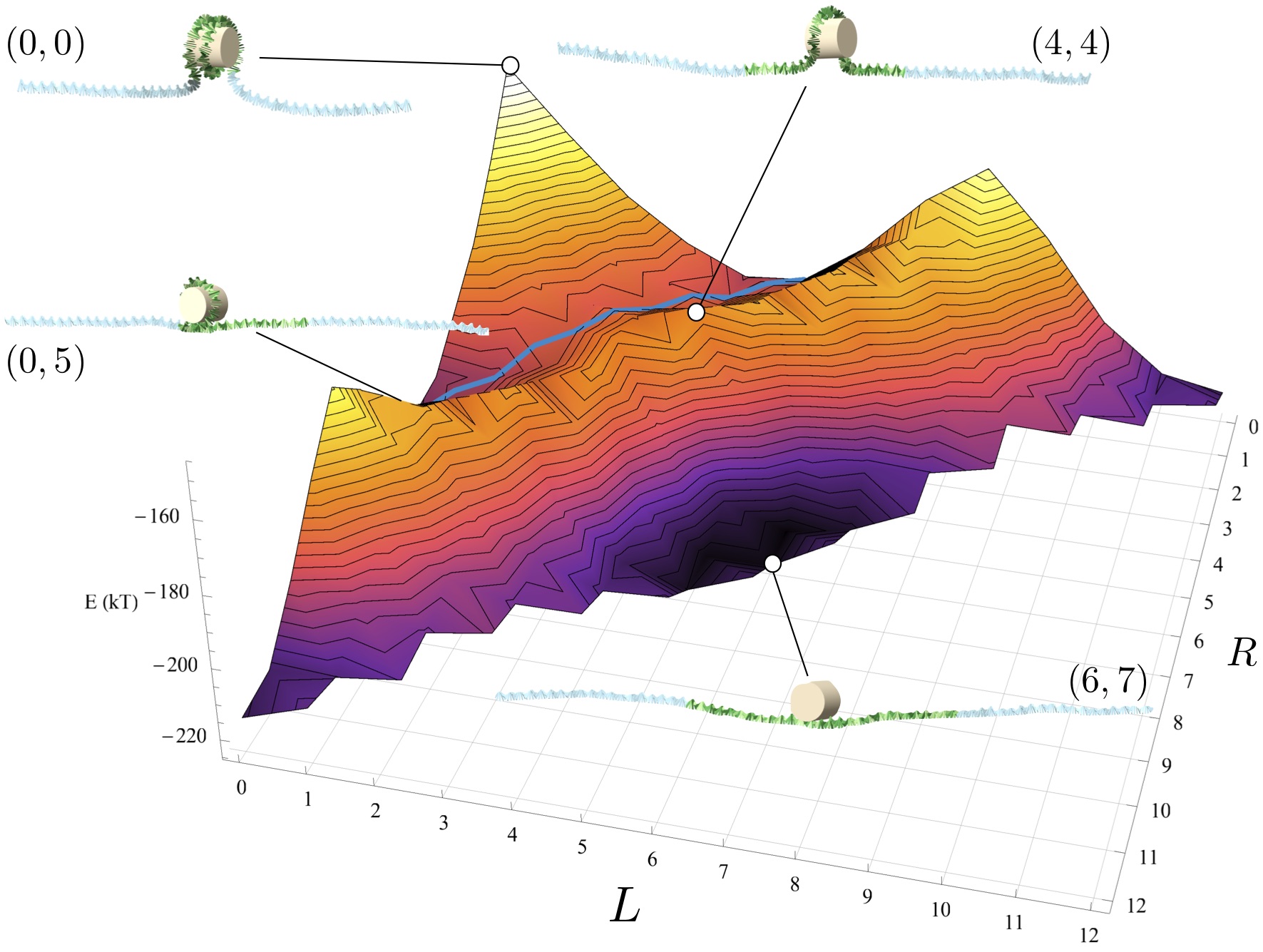
Another important question is how to explain the fact that nucleosomes seem to be much more stable
against external forces as one would expect based on other experiments (as e.g. the above mentioned
ones). This can be understood as a dynamical phenomenon: in order for a nucleosome
to unwrap under force, it has to flip by 180 degrees, severely bending its DNA. This
protects nucleosomes kinetically against transient external forces. Our computer model (de Bruin 2016)
can explain detailed experimental findings based on a combination of micromanipulation and FRET.
Using our Mutation Monte Carlo method we can also design special base-pair sequences
which lead to nucleosomes which unwrap encountering
hardly any barrier, constituting something like "force sensors" (Tompitak 2017).
For details check out:
J. Culkin, L. de Bruin, M. Tompitak, R. Phillips and H. Schiessel:
The role of DNA sequence in nucleosome breathing, Eur. Phys. J. E
40, 106 (2017)
K. van Deelen, H. Schiessel and L. de Bruin:
Ensembles of breathing nucleosomes: a computational study, Biophys. J., in press (2020)
L. de Bruin, M. Tompitak, B. Eslami-Mossallam and H. Schiessel:
Why Do Nucleosomes Unwrap Asymmetrically?, J. Phys. Chem. B
120, 5855-5863 (2016)
M. Tompitak, L. de Bruin, B. Eslami-Mossallam and H. Schiessel: Designing nucleosomal force sensors, Phys. Rev. E
95, 052402 (2017)