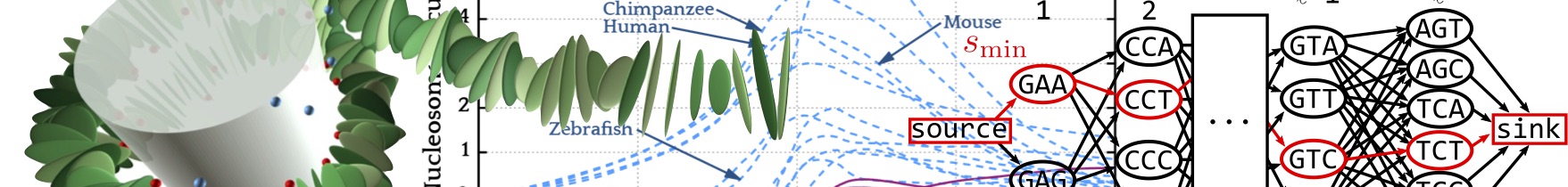
Despite being heavily coarse grained our
nucleosome model is much too slow to study the nucleosome energy landscape along entire
genomes. However, we have achieved a 10 million times speed-up, by using our
model to construct a probabilistic model, a two-step Markov process that assigns
to every nucleosomal base-pair sequence a free energy cost
(Tompitak 2017a; Neipel 2020).
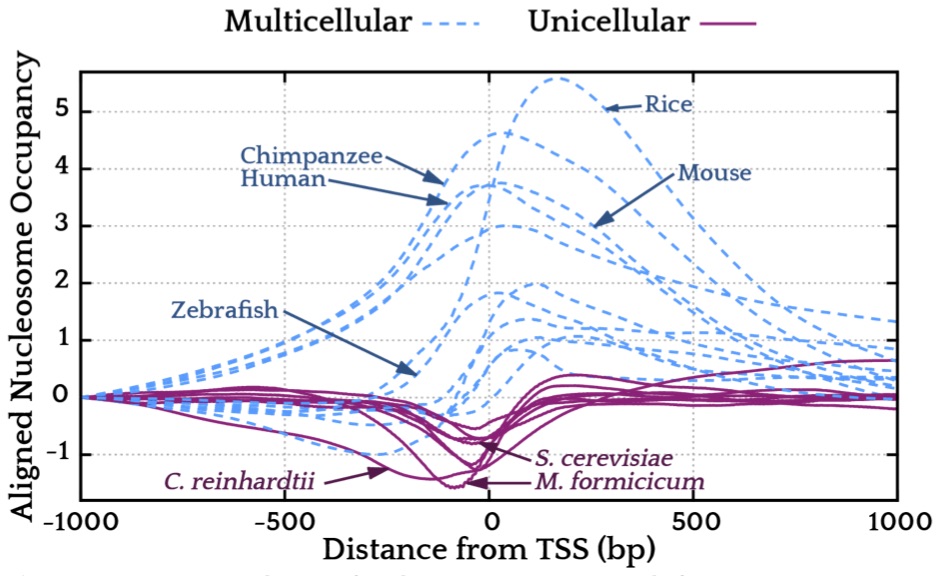
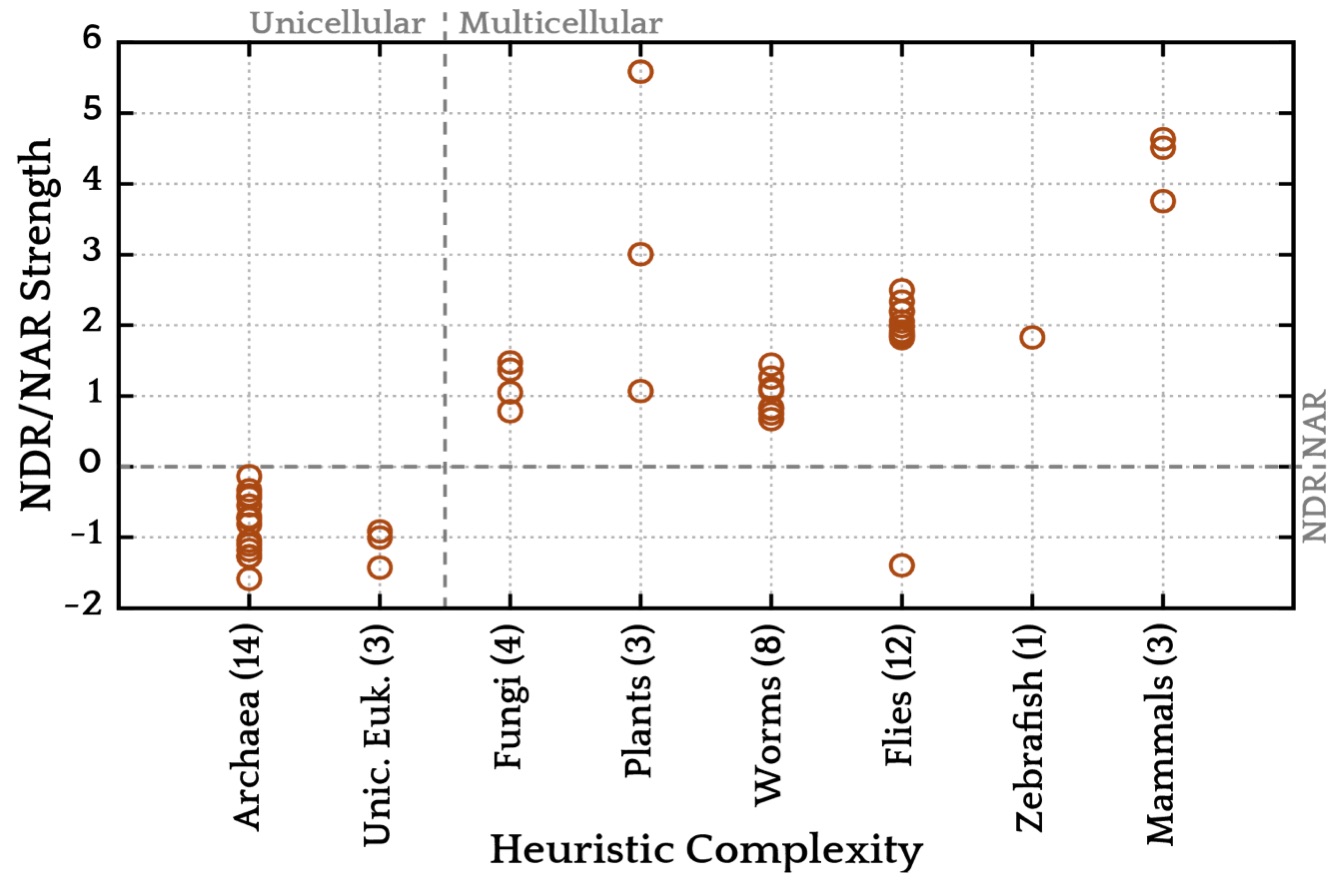
Using this probabilistic model we looked at the genome-wide average of the nucleosome free energy
landscape around transcription start sites for more than 50 genomes
(Tompitak 2017b). We found a general rule that
seems to apply to all living organisms: if the organism is unicellular, DNA mechanics
repels nucleosomes from that region and, if it is multicellular, it attracts nucleosomes
on average. The biological reason behind this distinction is probably rather complex
and might involve transfer of epigentic information from fathers to offsprings.
For details check out:
M. Tompitak, R. T. Barkema and H. Schiessel:
Benchmarking and refining probability-based models for nucleosome-DNA interaction, BMC Bioinformatics
18, 157 (2017a)
M. Tompitak, C. Vaillant and H. Schiessel:
Genomes of multicellular organisms have evolved to attract nucleosomes to promoter regions, Biophys. J.
112, 505-511 (2017b)
J. Neipel, G. Brandani and H. Schiessel:
Translational nucleosome positioning: a computational study, Phys. Rev. E
101, 022405 (2020)